With 25 years experience lecturing bioinformatics at university, I have progressively turned most of my teaching into hands-on problem solving using real world research datasets.
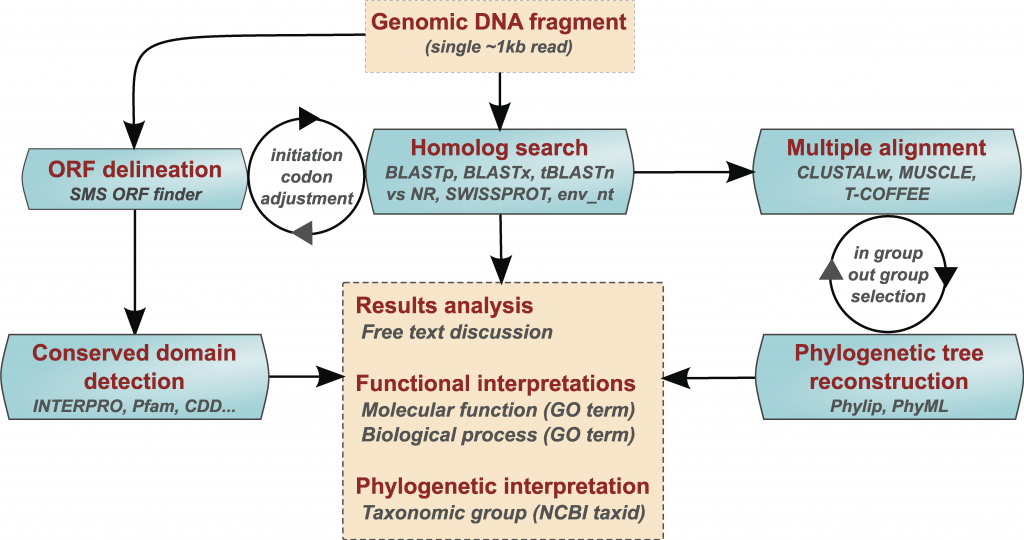
To support such a challenging approach to large student cohorts, I have developed a bioinformatics specific online e-learning environment called the Annotathon. In a flipped classroom setting, trainees are invited to annotate metagenomic DNA fragments added to their personal sequence cart from various environmental locations (such as those collected during the Tara Oceans Expedition, from human body sites of the Human Microbiome Project, or in 2021 a collection of coronavirus genomes). The analyses cover the fundamentals of sequence analysis, including the detection of potential coding regions, the search for sequence homologs and the identification of conserved protein domains, as well as phylogenetic trees inference.
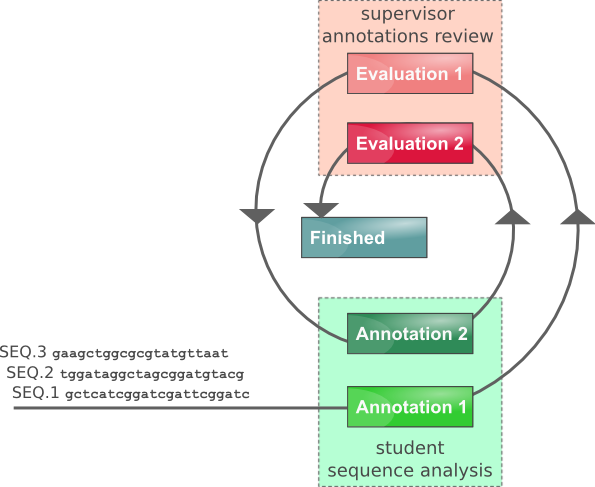
The Annotathon is designed to manage large trainee cohorts grouped into teams (such as students following the same course) supervised by instructors. The Annotathon serves as a numeric lab book, collecting trainee protocols, raw results, analyses and interpretations, collectively referred to as annotations. Each sequence annotation produced by trainees is examined, commented on and evaluated by instructors twice. This allows trainees to revise their initial annotations, thus improving their bioinformatics skills. After the team closing date, trainee evaluations are compiled into an overall grade.
Over 200 cohorts have been trained using the Annotathon since its inception in 2005, with teams from all continents joining in after the Annotathon was featured in the November 2008 edition of PLoS Biology: “Metagenome Annotation Using a Distributed Grid of Undergraduate Students“. I have also had the pleasure of teaching Annotathon based computational biology introductory courses in Brazzaville (Congo in 2009, 2011, 2012, 2013 & 2014), Hanoi (Vietnam in 2015 & 2018) and Koweit (2009).