Research interests
Computational biology / bioinformatics : environmental genomics (especially metagenomes and metabarcodes), high throughput sequence analysis, plankton biology and ecology, virology (large DNA viruses, coronaviruses).
Grants and Research Contracts
- Invited Associate Professor, Kyoto University (Japan, May-Jul 2015)
- French ANR / Investement of the Future contract OCEANOMICS (ANR-11-BTBR-0008, 425.000€, 2012-2019)
- European Union FP7 contract MicroB3 (FP7-OCEAN-2011 287589, 275.000€, 2012-2016)
- Sabbatical (18 months @ CNRS IGS/UPR2589 & INSERM U928, 2009-2010),
- European Union FP5 contract TEMBLOR DESPRAD (QLRI-CT-2001-00015, 260.000€, 2001-2004)
- Canadian Medical Research Council, 3 years postdoctoral grant (ranked 19th/190, 1996-1998)
- Royal Victoria Hospital Research Institute (Canada), 1 year postdoctoral grant (1995)
- Institute for Animal Health (GB), 3 years doctoral training grant (1991-1993)
- EU ERASMUS grant (INSA graduate final year 6 months training, 1991)
Skills
Bioinformatics: classical & high throughput sequence analysis (homology searches, functional annotation, phylogeny), classical & environmental genomics (assembly, metagenomes & metabarcodes analyses), data mining & visualisation (GIS, heatmaps, classification, co-occurence networks, numerical ecology).
IT: system admin (Linux installation & admin, Bash, Apache, Git, virtualization, containers), HPC & compute farms (SLURM / OAR), programming (PHP, Perl/Laravel, MySQL/PostgreSQL, HTML/Javascript/Bootstrap, Nextflow workflow management)
Experimental wet lab biology: field sampling (filtration, fixing, metadata collection), cell biology & virology (cultures, mouse KO, virus titration & cultures), molecular biology (cloning, sequencing, amplification, Northern/Southern blots), biochemistry (protein purification, HPLS/SDS PAGE chromatography, Western blots, in vitro enzymatic assays).
Projects examples
- Marine plakton metagenomics : Ocean Gene Atlas
- Design of degenerate PCR primers for the amplification of giant virus barcodes : MegaPrimer
Productivity
- Clarivate Web of Science stats (Jan 2022):
- Publications: 42
- h-index: 32
- Citing articles: 6,892 (wo self-citations)
- ORCID publication record
- Research Gate
- Google Scholar
- Research laboratory web page
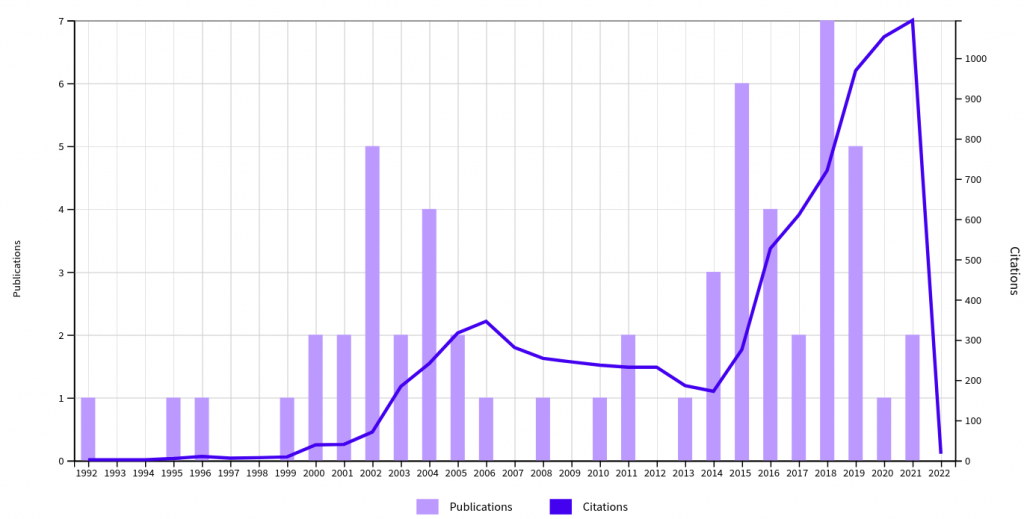
List of publications (selection with highest personal contributions)
- Diversity and evolution of bacterial bioluminescence genes in the global ocean. NAR Genomics and Bioinformatics, 2020 | journal-article DOI: 10.1093/nargab/lqaa018
- The Ocean Gene Atlas: exploring the biogeography of plankton genes online. Nucleic acids research, 2018 | journal-article DOI: 10.1093/nar/gky376
- Degenerate PCR primers to reveal the diversity of giant viruses in coastal waters. Viruses, 2018 | journal-article DOI: 10.3390/v10090496
- Linking virus genomes with host taxonomy. Viruses, 2016 | journal-article DOI: 10.3390/v8030066
- Ocean plankton. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science, 2015 | journal-article DOI: 10.1126/science.1261447
- Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. 2013 | journal-article DOI: 10.1038/ismej.2013.59
- Metagenome annotation using a distributed grid of undergraduate students. PLoS Biology, 2008 | journal-article DOI: 10.1371/journal.pbio.0060296
- MicroArray Facility: a laboratory information management system with extended support for Nylon based technologies. BMC Genomics, 2006 | journal-article DOI: 10.1186/1471-2164-7-240
- Mice lacking the UBC4-testis gene have a delay in postnatal testis development but normal spermatogenesis and fertility. Molecular & Cellular Biology, 2005 | journal-article DOI: 10.1128/MCB.25.15.6346-6354.2005
- Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics, 2001 | journal-article DOI: 10.1038/ng1201-365
- The EMBL Nucleotide Sequence Database. Contributing and accessing data. Molecular Biotechnology, 1999 | journal-article DOI: 10.1385/MB:12:3:255
- A novel rat homolog of the Saccharomyces cerevisiae ubiquitin- conjugating enzymes UBC4 and UBC5 with distinct biochemical features is induced during spermatogenesis. Molecular and Cellular Biology, 1996 | journal-article DOI: 10.1128/MCB.16.8.4064
- Characterization of a ubiquitinated protein which is externally located in African swine fever virions. Journal of Virology, 1995 | journal-article PMC: PMC188786
- A ubiquitin conjugating enzyme encoded by African swine fever virus. EMBO Journal, 1992 | journal-article PMC: PMC556456
List of publications (all)
- Deep ocean metagenomes provide insight into the metabolic architecture of bathypelagic microbial communities. Communications Biology, 2021 | journal-article DOI: 10.1038/s42003-021-02112-2
- The Ocean barcode atlas: A web service to explore the biodiversity and biogeography of marine organisms. Molecular Ecology Resources, 2021 | journal-article DOI: 10.1111/1755-0998.13322
- Diversity and evolution of bacterial bioluminescence genes in the global ocean. NAR Genomics and Bioinformatics, 2020 | journal-article DOI: 10.1093/nargab/lqaa018
- Microbial abundance, activity and population genomic profiling with mOTUs2. Nature Communications, 2019 | journal-article DOI: 10.1038/s41467-019-08844-4
- Gene Expression Changes and Community Turnover Differentially Shape the Global Ocean Metatranscriptome. Cell, 2019 | journal-article DOI: 10.1016/j.cell.2019.10.014
- Minimum Information about an Uncultivated Virus Genome (MIUViG). Nature Biotechnology, 2019 | journal-article DOI: 10.1038/nbt.4306
- Locality and diel cycling of viral production revealed by a 24 h time course cross-omics analysis in a coastal region of Japan. The ISME journal, 2018 | journal-article DOI: 10.1038/s41396-018-0052-x
- Taxon Richness of “Megaviridae” Exceeds those of Bacteria and Archaea in the Ocean. Microbes and environments, 2018 | journal-article DOI: 10.1264/jsme2.me17203
- The Ocean Gene Atlas: exploring the biogeography of plankton genes online. Nucleic acids research, 2018 | journal-article DOI: 10.1093/nar/gky376
- A global ocean atlas of eukaryotic genes. Nature communications, 2018 | journal-article DOI: 10.1038/s41467-017-02342-1
- Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nature communications, 2018 | journal-article DOI: 10.1038/s41467-017-02235-3
- Degenerate PCR primers to reveal the diversity of giant viruses in coastal waters. Viruses, 2018 | journal-article DOI: 10.3390/v10090496
- Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Scientific data, 2017 | journal-article DOI: 10.1038/sdata.2017.93
- Environmental Viral Genomes Shed New Light on Virus-Host Interactions in the Ocean. MSphere, 2017 | journal-article DOI: 10.1128/msphere.00359-16
- Partial gene catalog of deep samples (4000m) from a Global Metagenomics Expedition (Malaspina 2010). PANGAEA – Data Publisher for Earth & Environmental Science 2017 | data-set DOI: 10.1594/pangaea.883794
- Plankton networks driving carbon export in the oligotrophic ocean. Nature, 2016-02 | journal-article DOI: 10.1038/nature16942
- Cyanobacterial symbionts diverged in the late Cretaceous towards lineage-specific nitrogen fixation factories in single-celled phytoplankton. Nature Communications, 2016 | journal-article DOI: 10.1038/ncomms11071
- Linking virus genomes with host taxonomy. Viruses, 2016 | journal-article DOI: 10.3390/v8030066
- Reverse transcriptase genes are highly abundant and transcriptionally active in marine plankton assemblages. ISME Journal, 2016 | journal-article DOI: 10.1038/ismej.2015.192
- Deep sequencing of amplified Prasinovirus and host green algal genes from an Indian Ocean transect reveals interacting trophic dependencies and new genotypes. Environmental Microbiology Reports, 2015-12 | journal-article DOI: 10.1111/1758-2229.12345
- Ocean plankton. Determinants of community structure in the global plankton interactome. Science, 2015-05 | journal-article DOI: 10.1126/science.1262073
- Ocean plankton. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science, 2015-05 | journal-article DOI: 10.1126/science.1261447
- Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science, 2015-05 | journal-article DOI: 10.1126/science.1261605
- Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science, 2015-05 | journal-article DOI: 10.1126/science.1261498
- Ocean plankton. Structure and function of the global ocean microbiome. Science, 2015-05 | journal-article DOI: 10.1126/science.1261359
- Open science resources for the discovery and analysis of Tara Oceans data. Scientific Data, 2015 | journal-article DOI: 10.1038/sdata.2015.23
- QDD version 3.1: a user-friendly computer program for microsatellite selection and primer design revisited: experimental validation of variables determining genotyping success rate. Molecular Ecology Resources, 2014 | journal-article DOI: 10.1111/1755-0998.12271
- Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environmental Microbiology, 2014 | journal-article DOI: 10.1111/1462-2920.12250
- Unveiling of the diversity of Prasinoviruses (Phycodnaviridae) in marine samples by using high-throughput sequencing analyses of PCR-amplified DNA polymerase and major capsid protein genes. Applied and Environmental Microbiology, 2014 | journal-article DOI: 10.1128/AEM.00123-14
- Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. 2013-09 | journal-article DOI: 10.1038/ismej.2013.59
- A holistic approach to marine eco-systems biology. 2011-10 | journal-article DOI: 10.1371/journal.pbio.1001177
- Annotation distribuée, les étudiants ouvrent la voie. Biofutur 2011 | journal-article EID: 2-s2.0-80051773652
- mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. 2010-05 | journal-article DOI: 10.1101/gr.102582.109
- What bioinformatics can tell us about ourselves and our environment? Proceedings of the 2009 conference on Information Science, Technology and Applications – ISTA 2009 | conference-paper DOI: 10.1145/155195PMID: 20360389
- Metagenome annotation using a distributed grid of undergraduate students. PLoS Biology, 2008-11 | journal-article DOI: 10.1371/journal.pbio.0060296
- MicroArray Facility: a laboratory information management system with extended support for Nylon based technologies. BMC Genomics, 2006 | journal-article DOI: 10.1186/1471-2164-7-240
- Mice lacking the UBC4-testis gene have a delay in postnatal testis development but normal spermatogenesis and fertility. Molecular & Cellular Biology, 2005-08 | journal-article DOI: 10.1128/MCB.25.15.6346-6354.2005
- An open letter on microarray data from the MGED Society. Microbiology, 2004-11 | journal-article PMID: 15528642
- Submission of microarray data to public repositories. PLoS Biology, 2004-09 | journal-article DOI: 10.1371/journal.pbio.0020317
- Standards for microarray data: an open letter. Environmental Health Perspectives, 2004-08 | journal-article PMC: PMC1277123
- Feature extraction and signal processing for nylon DNA microarrays. BMC Genomics, 2004-06 | journal-article DOI: 10.1186/1471-2164-5-38
- DNA microarray data and contextual analysis of correlation graphs. BMC Bioinformatics, 2003-04 | journal-article DOI: 10.1186/1471-2105-4-15
- Breast cancer revisited using DNA array-based gene expression profiling. International Journal od Cancer, 2003-02 | journal-article DOI: 10.1002/ijc.10867
- Prognosis of breast cancer and gene expression profiling using DNA arrays. Annals of the NY Academy of Sciences, 2002-12 | journal-article DOI: 10.1111/j.1749-6632.2002.tb05954.x
- The underlying principles of scientific publication. Bioinformatics, 2002-11 | journal-article DOI: 10.1093/bioinformatics/18.11.1409
- Standards for microarray data. Science, 2002-10 | journal-article DOI: 10.1126/science.298.5593.539b
- EMBL-Align: a new public nucleotide and amino acid multiple sequence alignment database. Bioinformatics, 2002-05 | journal-article DOI: 10.1093/bioinformatics/18.5.763
- A guide to microarray experiments – An open letter to the scientific journals. Lancet, 2002 | journal-article DOI: 10.1016/S0140-6736(02)11100-7
- Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics, 2001-12 | journal-article DOI: 10.1038/ng1201-365
- Divergent N-Terminal Sequences Target an Inducible Testis Deubiquitinating Enzyme to Distinct Subcellular Structures. Molecular and Cellular Biology, 2001-02 | journal-article PMC: PMC86687
- The EMBL nucleotide sequence database. Nucleic Acids Research, 2000 | journal-article DOI: 10.1093/nar/28.1.19
- The EMBL Nucleotide Sequence Database. Contributing and accessing data. Molecular Biotechnology, 1999 | journal-article DOI: 10.1385/MB:12:3:255
- A novel rat homolog of the Saccharomyces cerevisiae ubiquitin- conjugating enzymes UBC4 and UBC5 with distinct biochemical features is induced during spermatogenesis. Molecular and Cellular Biology, 1996 | journal-article DOI: 10.1128/MCB.16.8.4064
- Characterization of a ubiquitinated protein which is externally located in African swine fever virions. Journal of Virology, 1995 | journal-article PMC: PMC188786
- African swine fever virus genome content and variability. Arch Virol, 1993 | journal-article PMID: 821980
- A ubiquitin conjugating enzyme encoded by African swine fever virus. EMBO Journal, 1992 | journal-article PMC: PMC556456