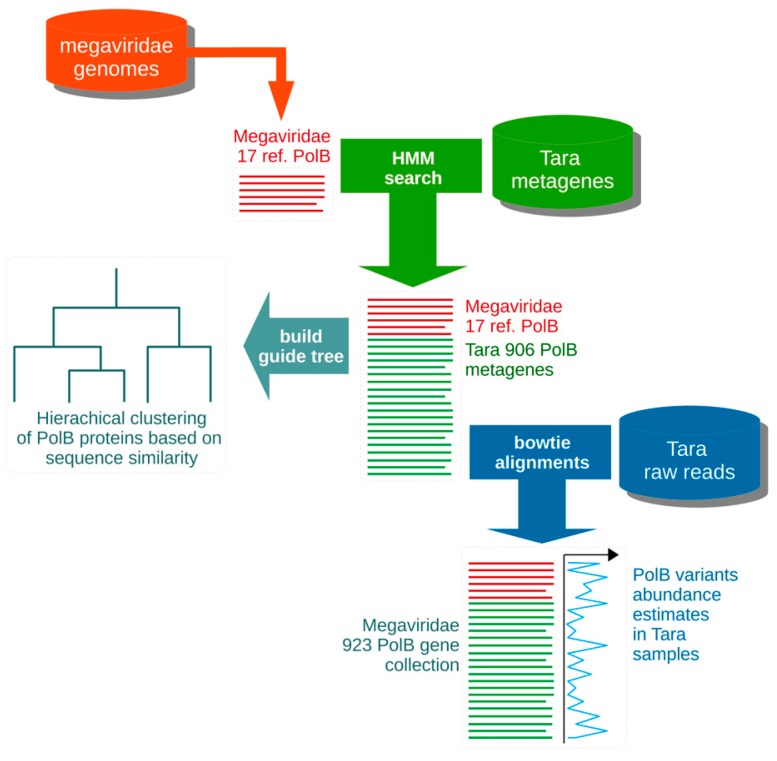
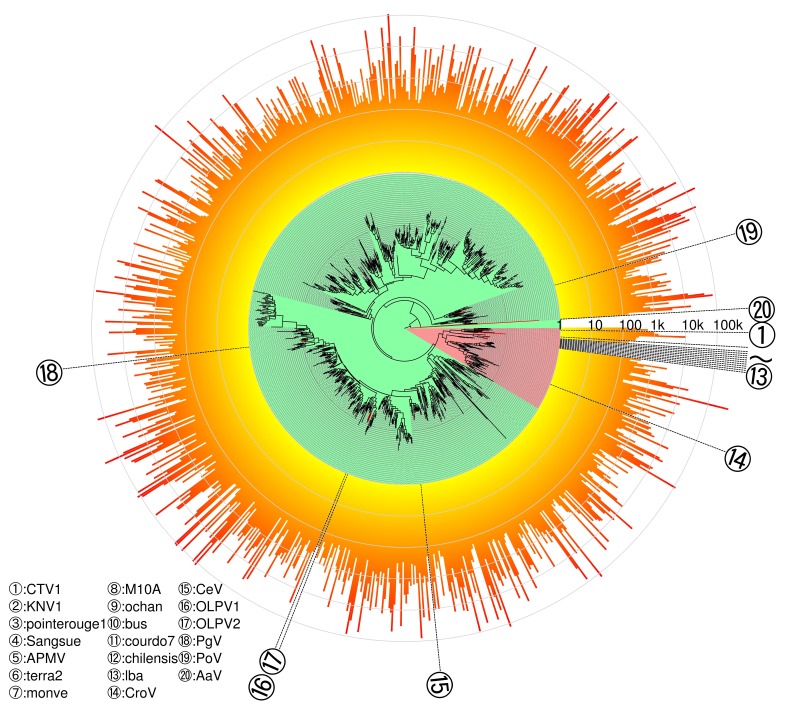
The diversity of giant double stranded viruses in the sea is extremely high, but they are notoriously difficult to observe and cultivate. In collaboration with my colleagues from Kyoto University, I developped a bioinformatics workflow to design a set of primers that target all known members of the megaviridae. The workflow, named UjiLity after the name of the Kyoto campus where I had the privilege to develop it, identifies the best conserved subregion of the target dataset verifying user defined constraints, including amplicon length, oligonucleotide length and precise Tm, as well as maximal allowed degeneracy. The specific challenge was to cater for the extreme divergence between the viral target sequences, by allowing multiple primer pairs to be defined with constrained degeneracy. The UjiLity scripts together with the giant virus input data is available on https://github.com/PHingamp/UjiLity.
The “MEGAPRIMER” set of oligos designed using the UjiLity workflow was tested with marine water samples and showed this method allowed much deeper surveys of giant viruses diversity in environmental samples. For more details, see Viruses 2018 “Degenerate PCR Primers to Reveal the Diversity of Giant Viruses in Coastal Waters”.